What are restriction enzymes?
Molecular scissors that cut double-stranded DNA molecules at specific sites. These are a unique class of enzymes that cleave (cut) DNA at a particular distinct interior place along with its size. Frequently called restriction endonucleases (Since they cut within the molecule).
Biological Function
A lot of bacteria use restriction enzymes as protection against bacteriophages. Restriction enzymes avoid the duplication of the phage by cleaving its DNA at certain sites. The host DNA is shielded by Methylases which add methyl groups to adenine or cytosine bases within the site thus changing the site and shielding the DNA.
History of Restriction Enzyme
The first restriction enzyme was isolated in 1970 by Hindll. He did the subsequent exploration and characterization of numerous restriction endonucleases. From after that, over 3000 restriction enzymes have actually been examined in detail, and more than 600 of these are readily available and are routinely made use of for DNA modification and manipulation in labs.
Nomenclature
Since their discovery in the 1970s, numerous restriction enzymes have actually been determined; for instance, more than 3500 different Type II restriction enzymes have been characterized.
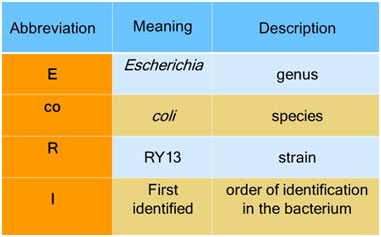
Each enzyme is called after the microorganism from which it was separated, making use of a naming system based upon bacterial genus, species, and strain. For example, the name of the Eco RI restriction enzyme was derived as shown:
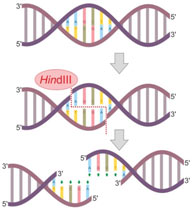
Mode of Action of Restriction Enzymes
The enzyme makes two lacerations, one with each of the sugar-phosphate backbone (i.e., each strand) of the double helix without damaging the nitrogenous bases. Restriction enzymes hydrolyze the backbone of DNA between deoxyribose and phosphate groups.
This leaves a phosphate group on the 5′ ends and hydroxyl on the 3′ ends of both strands. A few restriction enzymes will certainly cleave single-stranded DNA, although typically at low efficiency.
The restriction enzymes most made use of in molecular biology laboratories cut within their recognition sites and generate one of three various kinds of ends in the layouts listed below,
- 5′ overhangs: The enzyme cuts asymmetrically within the recognition site such that a brief single-stranded section prolongs from the 5′ end. BamHI cuts in this way.
- 3′ overhangs: The enzyme cuts asymmetrically within the recognition site such that the result is a single-stranded overhang from the two 3′ ends. KpnI cuts in this fashion.
- Blunts: Enzymes that cut at exactly opposite sites in both strands of DNA generate blunt ends without overhangs. SmaI is an instance of an enzyme that creates blunt.
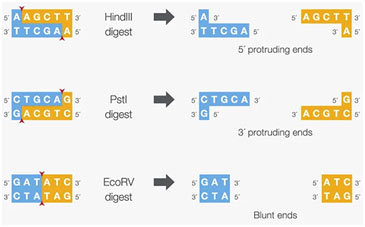
- Sticky ends
The 5′ or 3′ overhangs produced by enzymes that cut asymmetrically are called sticky ends or cohesive ends, since they will easily stick or anneal with their partner by base pairing. The sticky end is also called a cohesive end or complementary end.
Star Activity of Restriction Enzyme
Star activity is defined as the alteration in the digestion uniqueness that takes place under sub-optimal enzyme conditions. Star activity leads to cleavage of DNA at non-specific sites. Some of the suboptimal conditions that result in Star activity are as follows:
- pH >8.0
- glycerol concentration of >5%
- enzyme concentration >100 units/mg of DNA
- increased incubation time with the enzyme
- incorrect cofactor or buffer
The Influence of Restriction Enzymes
Genetic Engineering
- Type II enzymes produced many sensible benefits, like E. coli K12, its genetics, and its vectors became the workhorses of molecular biology in the 1970s for cloning, generation of libraries, DNA sequencing, discovery, and also an overflow of enzymes, hormones, etc.
- The applications of Type II enzymes remained to expand, especially after the arrival of synthetic DNA, in vitro product packaging of DNA in phage bits as well as enhanced bacterial hosts and vectors for overexpression and stabilization of proteins.
- Production of insulin from recombinant microorganisms and yeast, thus significantly enhancing the supply for diabetics and the manufacturing of a recombinant vaccine for Hepatitis B to deal with the thousands of millions of people at risk of infection by this virus.
- Restriction enzymes are devices for keeping track of RestrictionFragment Length Polymorphisms (RFLP), enabling the location of mutations, generation of human linkage maps, recognition of disease genes (such as sickle cell characteristic or Huntington disease), as well as last, yet not the very least, the DNA fingerprinting strategy created by Alec Jeffreys.
- DNA fingerprinting allows the solution of paternal cases, the identification of bad guys as well as their sufferers as well as the exoneration of the wrongly charged.
- Making use of REases in this system enabled the development of appropriate treatments for such identification.
- Common restriction enzymes are used in cloning experiments to recognize four-, six-, or eight-base series.